Exercise 3: Barbituric Acid Dihydrate
Three Unique Molecules
Initial Analysis
- Open the BARBAD01 CIF

- Complete all molecules

- For each molecule:
- Generate Hirshfeld surfaces
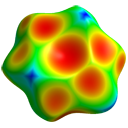
- Map with HF/3-21G electrostatic potential
- Generate Hirshfeld surfaces
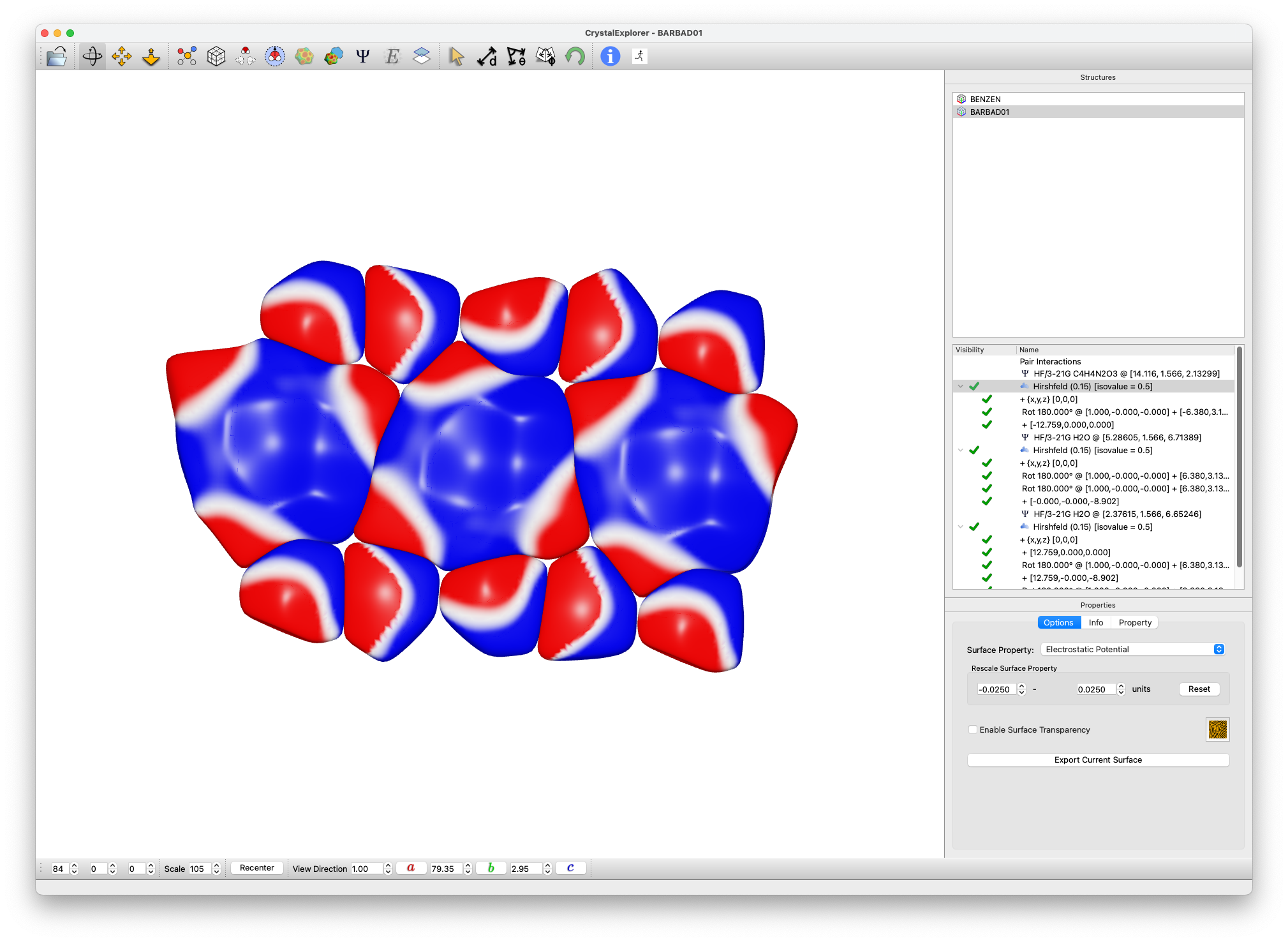
Surface Analysis
- For each Hirshfeld surface:
- Rescale surface property to ±0.025 au range
- Note the planar arrangement of the cluster
- Extend to create a sheet of hydrogen-bonded molecules using Generate External Fragment
- Use Clone Surface
 for each molecule
for each molecule
tip
Clone surfaces separately for each parent surface in the right-hand dialog
info
Observe the persistent pattern of electrostatic complementarity between adjacent molecules
Energy Analysis
- For each molecule:
- Select Reset Crystal between calculations
- Select the molecule
- Create 3.8 Å cluster
- Calculate energies
Energy Framework Analysis
- Generate unit cell molecules

- Create energy frameworks (Display / Energy frameworks):
- Compare:
- Coulomb Energy ()
- Dispersion Energy ()
- Total Energy ()
- Observe:
- Strong interactions between molecules in layers (especially )
- Weaker inter-layer interactions (mainly )
- Compare:
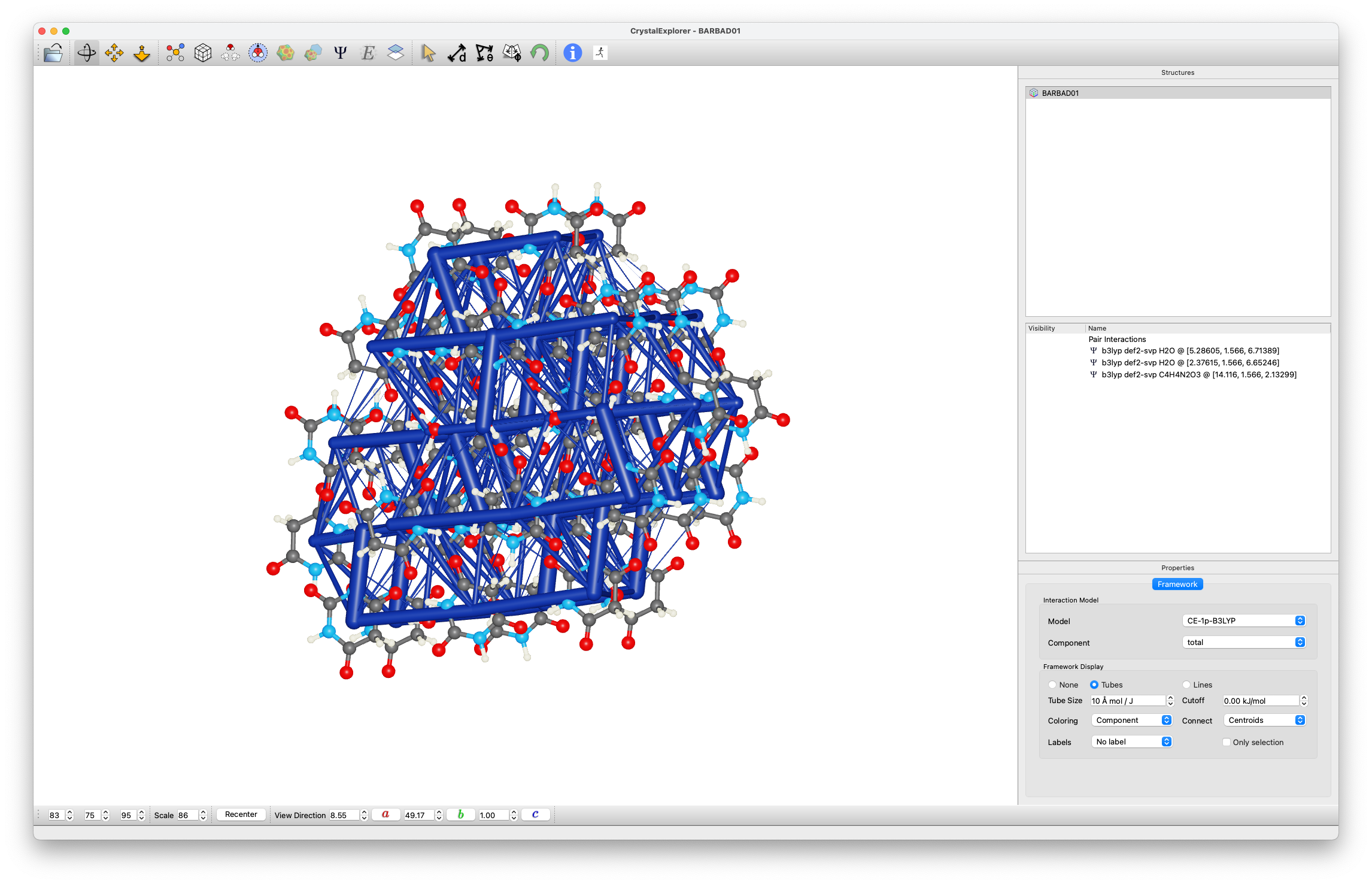
Historical Context
These results help explain the observation "There is marked cleavage parallel to (010)" from the original structure determination by Jeffrey et al.